
" Three points (atoms) common to the two structures were considered as the points of superimposition, and the distances between them were set to 0.001 k. The optimized geometries were superimposed using the overlay command in Chem3D Ultra 8.0 from CambridgeSoft, Cambridge, Massachusetts.
#Chemdraw ultra 3d view software#
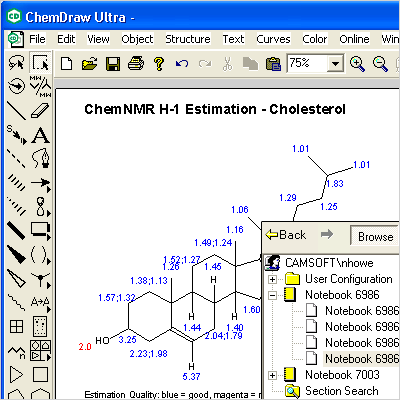
Interactions between template and monomer in MIPs were analyzed using Amber MM method 3-D chemical structures of the labeled BLAs were modeled using molecular mechanics using (MOPAC, AMI force field) using Chem3D Ultra 7.0 software Energy minimization by molecular mechanics and quantum chemistry to estimate enthalpies of formation, bond orders, intermolecular distances and ionization potentials using PCModel for windows. 7.0.0 by CambridgeSoft.ĬS Chem3D Ultra Cambridge Soft Corporation, Cambridge, USA. All 3D structures in this and the following figures are MM2 minimized in CS Chem3D Ultra v. 9.5 Schlegel diagrams and 3D structures of 1,2-CmII2 (la) and 1,4-CmH, (lb). (Printed with permission of CambridgeSoft.įig. Visualized using Cambridge Soft Chem3D Ultra 10.0 with notations in ChemDraw Ultra 10.0. įigure 2.10 Secondary and tertiary structure of the enzyme lysozyme, PDB 2C80.
#Chemdraw ultra 3d view full#
E-Notebook Ultra 7.0, BioAssay Pro 7.0, MOPAC, Gaussian GAMESS interfaces, ChemSAR Server Excel, CLogP, Purchasing for Excel, CombiChem/Excel, as well as the full set of Chemlnfo databases, including ChemACX ChemACX-SC, The Merck Index and ChemMSDX have been added to ChemOffice Pro. ĬhemDraw Ultra 7.0, Chem3D Ultra 7.0, and ChemFinder Pro 7.0 became available in 2002. Name/ version SymApps Chem3D Ultra 7.0 ACD/3D View UltraMol 2003 Rasmol Molmol JMol chime 2.63 Ortep III VieiverLite.


 0 kommentar(er)
0 kommentar(er)
